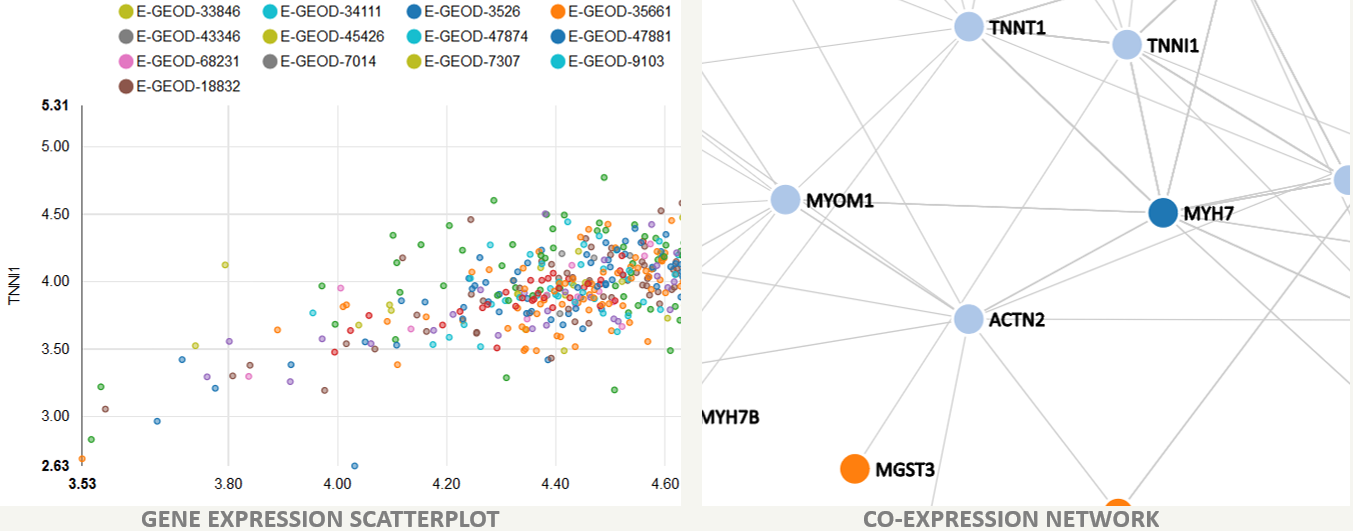
We have gathered the last 10 years of muscle-related microarray data from the public repository Gene Expression Omnibus (GEO) and Array Express (AE) to perform correlation analyses on both human and mouse organisms. Following searches with muscle-related keywords, the details of specific studies and samples were screened manually to select only those samples pertinent to muscle research.
Co-expression analyses will help muscle researchers to delineate the tissue-, cell-, and pathology-specific elements of muscle protein interactions, cell signaling and gene regulation. Changes in co-expression between pathologic and healthy tissue may suggest new disease mechanisms and therapeutic targets. MyoMiner is a powerful muscle specific database for the discovery of genes that are associated in related functions based on their co-expression.
To access the tool press Search on the Menu bar.